1. Target Prediction
1) Download the external validation dataset used in Target Prediction
Download external validation dataset click here!
Download external validation dataset click here!
Table 5. Full name of targets included in bioactivity prediction
| Abbreviations | Target name |
|---|---|
| 11β-HSD1 | 11β-hydroxysteroid dehydrogenase type 1 |
| AKT1 | Serine/threonine-protein kinase AKT1 |
| AKT2 | Serine/threonine-protein kinase AKT2 |
| AKT3 | Serine/threonine-protein kinase AKT3 |
| ALK | Anaplastic lymphoma kinase |
| ACE | Angiotensin-converting enzyme |
| AURKA | Aurora kinase A |
| AURKB | Aurora kinase B |
| BCL2 | Apoptosis regulator BCL-2 |
| BRAF | Serine/threonine-protein kinase B-raf |
| BRD4 | Bromodomain containing 4 |
| BTK | Bruton's tyrosine kinase |
| CCR2 | C-C chemokine receptor type 2 |
| c-Src | C-src tyrosine kinase |
| CDK1 | Cyclin dependent kinase 1 |
| CDK2 | Cyclin dependent kinase 2 |
| CHK1 | Checkpoint kinase 1 |
| CB1R | Cannabinoid receptor 1 |
| Cathepsin B | Cathepsin B |
| Cathepsin K | Cathepsin K |
| Cathepsin S | Cathepsin S |
| DPP-4 | Dipeptidyl peptidase 4 |
| EGFR | Epidermal growth factor receptor |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FGFR2 | Fibroblast growth factor receptor 2 |
| FGFR3 | Fibroblast growth factor receptor 3 |
| FLT3 | FMS-like tyrosine kinase 3 |
| HDAC1 | Histone deacetylase 1 |
| HDAC2 | Histone deacetylase 2 |
| HDAC6 | Histone deacetylase 6 |
| IGF1R | Insulin-like growth factor 1 receptor |
| JAK1 | Janus kinase 1 |
| JAK2 | Janus kinase 2 |
| JAK3 | Janus kinase 3 |
| MEK1 | Mitogen-activated protein kinase kinase 1 |
| MMP-2 | Matrix metallopeptidase 2 |
| MMP-3 | Matrix metallopeptidase 3 |
| MMP-9 | Matrix metallopeptidase 9 |
| MMP-13 | Matrix metallopeptidase 13 |
| MMP-14 | Matrix metallopeptidase 14 |
| MR | Mineralocorticoid receptor |
| NAMPT | Nicotinamide phosphoribosyl transferase |
| PDGFR-α | Platelet-derived growth factor receptor alpha |
| PDGFR-β | Platelet-derived growth factor receptor beta |
| PI3K-α | PI3-kinase p110-alpha subunit |
| PI3K-β | PI3-kinase p110-beta subunit |
| PI3K-δ | PI3-kinase p110-delta subunit |
| PI3K-γ | PI3-kinase p110-gamma subunit |
| PKC-θ | Protein kinase C theta |
| Renin | Renin |
| SYK | Spleen tyrosine kinase |
| TNF-α | Tumor necrosis factor-alpha |
| VEGFR1 | Vascular endothelial growth factor receptor 1 |
| VEGFR2 | Vascular endothelial growth factor receptor 2 |
| VEGFR3 | Vascular endothelial growth factor receptor 3 |
| ZAP70 | Tyrosine-protein kinase ZAP-70 |
Table 6. Description of ADMET endpoints
| Category | Property | Descriptions | Results Interpretation |
|---|---|---|---|
| Physicochemical Propertiy | MW | The measure of the sum of the atomic weight values of the atoms in a molecule | Molecular Weight |
| LogD7.4 | LogD7.4 is the 10-based logarithmic value of the octanol/water distribution coefficient at pH=7.4 and is used to measure the solubility of a compound in a lipid environment | The ideal range for LogD7.4 is 1~3 log mol/L | |
| LogS | LogS is the 10-based logarithmic of the aqeuous solubility value and is used to measure the ability of a compound to dissolve in water | The ideal range for LogS is -4 to 0.5 log mol/L | |
| LogP | LogP is the 10-based logarithmic value of octanol/water partition coefficient and is used to adduce how a drug molecule partitions itself into the lipid surroundings of the receptor microenvironment | The ideal range for TPSA is 0 to 3 log mol/L | |
| TPSA | TPSA(topological polar surface area) is the surface sum over all polar atoms and is used to measure the ability of a compound to penetrate cell membranes | The ideal range for TPSA is 0 to 140 | |
| Medicinal Chemistry | QED | QED(quantitative estimate of drug-likeness) is a method to quantify drug similarity as a value between 0 and 1 | The range is from 0 (unfavorable properties) to 1 (favorable properties) |
| SAscore | SA score (Synthetic Accessibility score) is a method to estimate the ease of synthesis of a compound with a value between 1 and 10 | The predicted value closer to 1 indicating that the compound is easy to synthesize, and closer to 10 indicating that the compound is difficult to synthesize | |
| Absorption | HIA | Human intestinal absorption (HIA) of oral drugs is critical for drug delivery to the target, and a molecule with less than 30% HIA is considered poorly absorbed | Category 0: HIA >=30%, Category 1: HIA <30%, and the output is the probability value belonging to Category 1, ranging from 0 to 1 |
| Pgp inhibitor | The inhibitor of P-glycoprotein. P-glycoprotein (Pgp) is an ABC transporter protein involved in intestinal absorption, drug metabolism, and brain penetration, and its inhibition can seriously alter a drug's bioavailability and safety | Category 0: Non-inhibitor, Category 1: Inhibitor, and the output is the probability value of being Pgp-inhibitor, ranging from 0 to 1 | |
| Distribution | BBB Penetration | As a membrane separating circulating blood and brain extracellular fluid, the blood-brain barrier (BBB) is the protection layer that blocks most foreign drugs, BBBP determines whether a compound can enter the brain region | Category 0: BBB- (logBB < -1), Category 1: BBB+ (logBB ≥ -1), and the output is the probability value of belonging to Category 1, ranging from 0 to 1 |
| Metabolism | CYP2C9 inhibitor | Cytochromes P450 (CYPs) are crucial in the metabolism of various molecules and chemicals within cells and contain 57 isoenzymes that metabolize approximately 2/3 of known drugs in humans, 80% of which are metabolized by 5 isozymes 1A2, 3A4, 2C9, 2C19 and 2D6. If a compound is a substrate for CYPs, it may be oxidized by enzymes, leading to inactive and/or toxic products, and if a compound is a CYP inhibitor, it may increase the concentration of other drugs, leading to drug interaction mediated toxicity | Category 0: Non-inhibitor(Non-substrate), Category 1: Inhibitor(Substrate), and the output is the probability value of being inhibitor(Substrate), ranging from 0 to 1 |
| CYP2D6 inhibitor | |||
| CYP3A4 inhibitor | |||
| CYP2C19 inhibitor | |||
| CYP1A2 inhibitor | |||
| CYP2D6 substrate | |||
| CYP3A4 substrate | |||
| Excretion | T1/2 | Half life(T1/2) of a drug is the duration for the concentration of the drug in the body to be reduced by half. When a drug has a short T1/2 (<3h), it usually indicates that the drug has a narrow therapeutic window and high toxicity | Category 0: T1/2 <3h, Category 1: T1/2 ≥3h, and the output is the probability value belonging to Category 1, ranging from 0 to 1. |
| Toxicity | Tox21 | Qualitative toxicity measurements on 12 biological targets, including nuclear receptors and stress response pathways, specifically NR-AR, NR-AR-LBD, NR-AhR, NR-Aromatase, NR-ER, NR-ER-LBD, NR-PPAR-gamma, SR-ARE, SR-ATAD5, SR-HSE, SR-MMP, SR-p53 | For each biological target, Category 0: inactives, Category 1: active, and the output is the probability value of being actives, ranging from 0 to 1 |
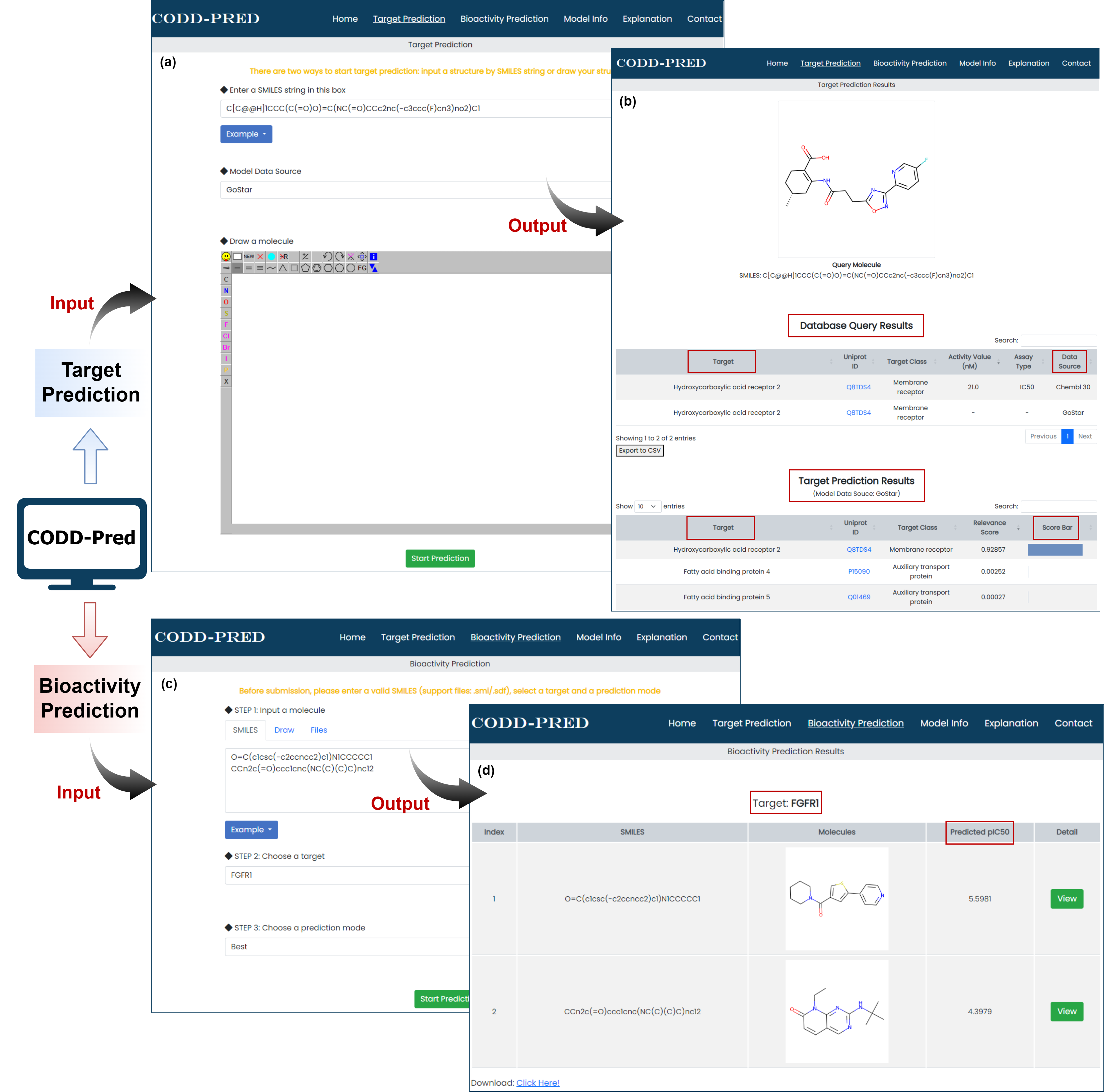
Figure 3. Graphical interface for input and output of CODD-PRED
Table 7. The development environment of CODD-PRED
| Library | Version |
|---|---|
| rdkit | 2020.09.1 |
| flask | 2.0.2 |
| pyg | 2.0.2 |
| dgl | 0.8.0 |
| dgllife | 0.2.9 |
| pytorch | 1.10.0 |
| torchvision | 0.7.0 |
| scikit-learn | 1.0.2 |
| xgboost | 1.5.1 |
| chemprop | 1.5.2 |
Table 8. Open source library license for website development
| Library | Author | License |
|---|---|---|
| chemprop | Wengong Jin et al. | MIT License |
| TDC | TDC Team | MIT License |
CODD-Pred: A Web Server for Efficient Target Identification and Bioactivity Prediction of Small Molecules. J. Chem. Inf. Model. 2023, 63, 20, 6169–6176.